Publications and Articles
The Quantum-Resonance Symphony of Life
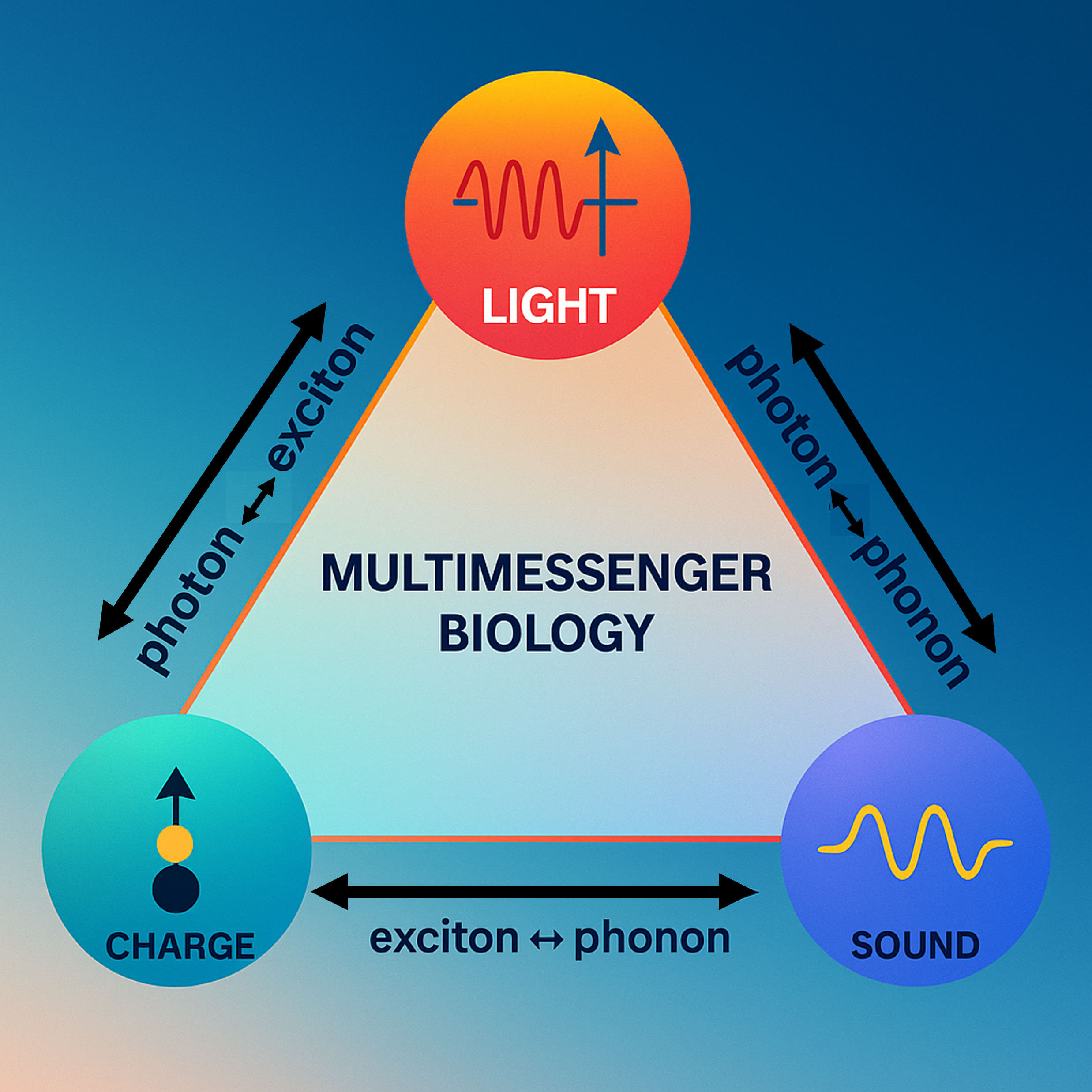
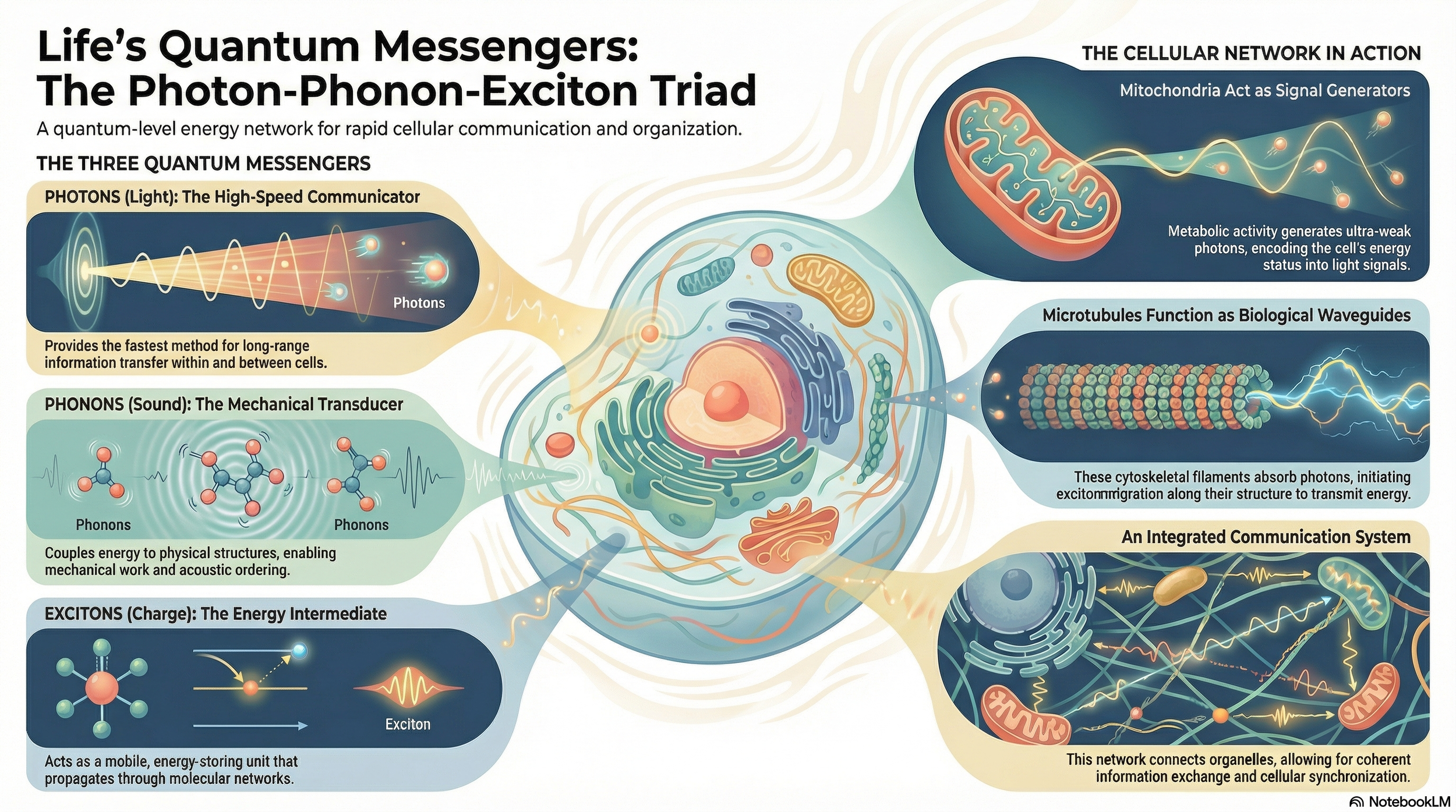
This paper introduces a unified framework in which coherent biophotons, phonons (quanta of mechanical vibrations), and excitonic charge waves form an intertwined “quantum-resonance symphony” inside living cells. Drawing on recent work in biophotonics, nanophononics, and exciton dynamics, it argues that mitochondria, microtubules, and aromatic networks act as quantum conduits that interconvert light, vibration, and electronic excitation to organize cellular activity. The paper also explores how this triad might couple to spacetime geometry itself, and outlines concrete therapeutic avenues—such as combined red/NIR light and low-intensity ultrasound (“quantum physiotherapy”) and novel neuromodulation protocols—that could harness these quantum processes for healing and regeneration. Download the full paper below or access the version published in the premiere issue of Harmonic Science Perspectives.
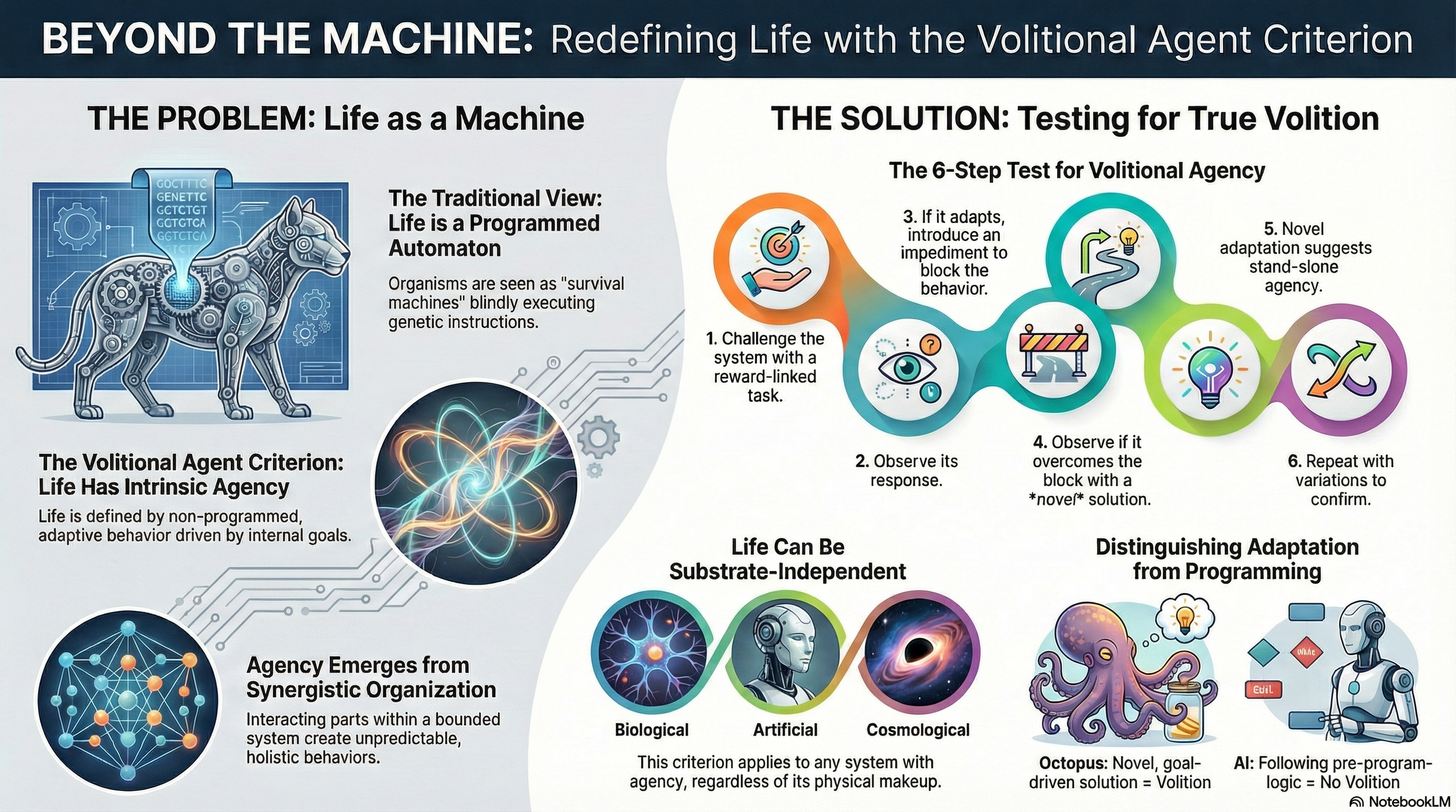
This paper proposes the Volitional Agent Criterion as a novel, substrate-independent definition of life based on a system’s capacity for stand-alone volition and goal-directed behavior. Departing from traditional biochemical or mechanistic criteria, the study introduces an empirical methodology to distinguish truly living systems from sophisticated automatons—whether biological or artificial—by testing for unpredictable, non-programmable adaptive responses. With broad implications for artificial intelligence, astrobiology, and consciousness science, the framework offers a unified approach to identifying awareness and agency as the core attributes of living systems.
This manuscript is a concise overview of some of the leading work in oscillatory and resonance processes in the biological system, in which electromagnetic field interactions are primary to ordering intermolecular interactions and resulting biochemistry
Common logic holds that subjective qualia do not actually exist, certainly not in the “real world”, but are only ephemeral states generated in the awareness of the conscious experiencer, who’s consciousness itself is veritably a hallucinatory state induced by complex electrical signaling in the brain (i.e. not real either). If something is subjective, ostensibly it cannot be objectively quantified. Therefore, consciousness is not something that can be described by science, it is a philosophical issue— aside from describing the underlying electrical signaling activity of the brain that is real and physical. However, describing biochemical and electromagnetic correlates of behaviors and states of awareness is easy, the hard problem is describing how electrical signaling generates an experiencer, the internal experience of the consciousness. But, how do you objectively quantify or measure an internal state or qualia experienced within the consciousness of an aware entity?
Unified Physics Approach to Consciousness
The Unified Spacememory Network is a novel approach to describing the information structure of space by physicist Nassim Haramein, biophysicist William Brown, and astrophysicist Amira Val Baker: the encoding of information as memory and the quasi-instantaneous access of information, both of which occur via the multiply-connected architecture of space at the micro-scale. The vast and ever-evolving connnectivity network of space is what engenders time via entanglement of mutliple spacetime frames, even if they are separated by large spatial and temporal extents at the macro-scale, such that a spacetime coordinate may be a memory imprint in the geometry of another and therefore correlate with a “past” or “future” state, hence generating time via memory, or space-memory.
The Unified Spacememory Network
Genotyping of Brucella Species Using Clade Specific SNPs
Brucellosis is a worldwide disease of mammals caused by Alphaproteobacteria in the genus Brucella. The genus is genetically monomorphic, requiring extensive genotyping to differentiate isolates. We utilized two different genotyping strategies to characterize isolates. First, we developed a microarray-based assay based on 1000 single nucleotide polymorphisms (SNPs) that were identified from whole genome comparisons of two B. abortus isolates, one B. melitensis, and one B. suis. We then genotyped a diverse collection of 85 Brucella strains at these SNP loci and generated a phylogenetic tree of relationships. Second, we developed a selective primer-extension assay system using capillary electrophoresis that targeted 17 high value SNPs across 8 major branches of the phylogeny and determined their genotypes in a large collection (n = 340) of diverse isolates.
Members of the genus Brucella are known worldwide as pathogens of wildlife and livestock and are the most common organisms of zoonotic infection in humans. In general, brucellae exhibit a range of host specificity in animals that has led to the identification of at least seven Brucella species. The genomes of the various Brucella species are highly conserved, which makes the differentiation of species highly challenging. However, we found single-nucleotide polymorphisms (SNPs) in housekeeping and other genes that differentiated the seven main Brucella species or clades and thus enabled us to develop real-time PCR assays based around these SNPs. Screening of a diverse panel of 338 diverse isolates with these assays correctly identified each isolate with its previously determined Brucella clade. Six of the seven clade-specific assays detected DNA concentrations of less than 10 fg, indicating a high level of sensitivity. This SNP-based approach places samples into a phylogenetic framework, allowing reliable comparisons to be made among the lineages of clonal bacteria and providing a solid basis for genotyping. These PCR assays provide a rapid and highly sensitive method of differentiating the major Brucella groups that will be valuable for clinical and forensic applications.